Abstract
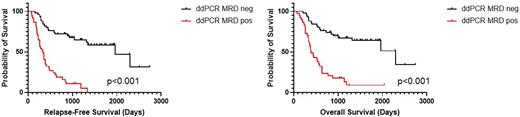
Measurable residual disease (MRD) is the most important post-treatment predictor of relapse in acute myeloid leukemia (AML) [PMID 33030517]. The bulk of clinical MRD studies in AML have been performed via multi-parameter flow cytometry and no molecular MRD platform is established for clinical use in AML. Droplet digital PCR (ddPCR) has been identified as a highly sensitive and precise modality for mutation monitoring in AML and other malignancies, but to date the range of mutations evaluated for AML has been limited [PMIDs 32629801, 28525762, 29472349]. Here we demonstrate the feasibility of retrospectively detecting molecular MRD with a broad panel of ddPCR assays in adult AML patients. We confirm the association of ddPCR MRD status with outcomes in patients receiving both standard chemotherapy and venetoclax/azacitidine, including a subset of patients who proceeded to stem cell transplant (SCT). All patients signed Colorado Multiple Institution Review Board (IRB) approved consent for collection of tissue used in this analysis, and an additional IRB approval allowed retrospective data analysis. Adult patients with a known AML mutational profile at diagnosis from clinical targeted NGS and with at least one post-remission bone marrow sample available for DNA extraction were included. Genomic DNA was extracted from whole bone marrow aspirates using Qiagen QIAsymphony DSP DNA kits. Concentration and quality of DNA was evaluated via Qubit spectrophotometer. A total of 57 AML-associated mutations were evaluated in this patient cohort using mutation-specific primer/probe ddPCR assays purchased commercially (BioRad) or custom designed. All assays were experimentally validated to a limit of detection of 0.02-0.15% variant allelic frequency (VAF). ddPCR was performed on a BioRad QX200 Droplet Digital PCR instrument using gDNA input of 150 ng per sample and appropriate positive and negative controls. Data were analyzed via the BioRad QuantaSoft Analysis Pro v1.0 software. Between one and five non-DTA mutations [PMID 29601269] per patient were serially monitored. For survival analyses, patients were categorized as MRD positive or negative according to their best response to therapy. Relapse-free survival (RFS) and overall survival (OS) were defined from the date of diagnosis to the respective endpoint. Individuals were censored at date of last follow-up. A total of 87 patients with at least one non-DTA mutation identified on diagnostic clinical NGS and available DNA from post-treatment time points had MRD quantified by ddPCR. Twenty patients received anthracycline + high-dose cytarabine-based therapies ('7+3'), 65 patients received venetoclax + azacitidine (ven/aza), and 2 patients received alternate low-intensity therapies (ivosedenib or decitabine + anti-CD123 antibody). Median follow-up for the entire cohort was 25.4 months (2.5-90.1 months). Fifty of 87 patients (19 '7+3', 30 ven/aza, 1 other) became MRD negative by ddPCR during the course of their therapy with median cohort survival of 36.4 months (6.9-90.1 months). The other 37 patients (1 '7+3', 35 ven/aza, 1 other) remained MRD positive at all evaluated time points post-therapy with median cohort survival of 12.9 months (2.5-67.1 months). Survival curves based on ddPCR MRD status are shown in Figure 1. A total of 39 patients (20 '7+3', 18 ven/aza, 1 "other" therapy) proceeded to SCT as part of their AML therapy, of which 9 were MRD negative prior to SCT, 27 became MRD negative post-SCT, and 3 had persistent MRD. There was no relationship between clearance of a mutation and its VAF at diagnosis (p=0.15). Of note, 12 patients (6 receiving '7+3', 6 receiving ven/aza) who achieved MRD negativity subsequently relapsed; of these 4 patients had reappearance of MRD by ddPCR between 1-4 months prior to clinical relapse. One patient was MRD negative pre-SCT and did not have post-SCT samples available prior to relapse. The other 7 patients had bone marrow evaluations every 2-3 months yet did not have a positive MRD time point prior to clinical relapse. ddPCR MRD is prognostic of outcomes in adult AML patients on therapy, including those receiving venetoclax/azacitidine. Evaluation of a broad range of mutations with ddPCR is possible and based on these data is expected to be a valuable prospective tool for clinical decisions about escalation or de-escalation of therapy.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal